| Model Browser User's Guide |
  |
Improving the Design
You can further optimize the design by returning to the Optimal Design dialog, where you can delete or add points optimally or at random. The most efficient way is to delete points optimally and add new points randomly -- these are the default algorithm settings. Only the existing points need to be searched for the most optimal ones to delete (the least useful), but the entire candidate set has to be searched for points to add optimally.
To strengthen the current optimal design:
- Return to the Design Editor window.
- Click the Optimal Design button in the toolbar again to reenter the dialog, and add 60 more points. Keep the existing points (which is the default).
- Click OK and watch the optimization progress, then click Accept when the number of iterations without improvement starts increasing.
- View the improvements to the design in the main displays.
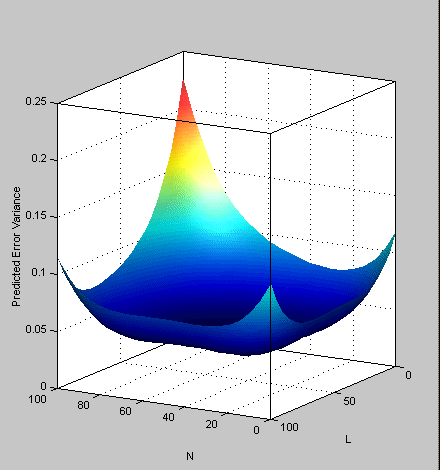
- Once again select Tools -> PEV Viewer and review the plots of prediction error variance and the new values of optimality criteria in the optimality frame (bottom left). The shape of the PEV projection might not change dramatically, but note the changes in the scales as the design improves. The values of D, V, and G optimality criteria will also change (you have to click Calculate to see the values).
To see more dramatic changes to the design, return to the Design Editor window (no need to close the PEV viewer).
- Split the display so you can see a 3-D projection at the same time as a Table view.
- Choose Edit -> Delete Point.
- Using the Table and 3-D views as a guide, in the Delete Points dialog, pick six points to remove along one corner; for example, pick points where N is 100 and L is 0. Add the relevant point numbers to the delete list by clicking the add (>) button.
- Click OK to remove the points. See the changes in the main design displays and look at the new Surface plot in the PEV viewer (see the example following).
 | Prediction Error Variance Viewer | | Classical Designs |  |





