

| Fuzzy Logic Toolbox |   |
Subtractive Clustering
Suppose we don't have a clear idea how many clusters there should be for a given set of data. Subtractive clustering, [Chi94], is a fast, one-pass algorithm for estimating the number of clusters and the cluster centers in a set of data. The cluster estimates obtained from the subclust function can be used to initialize iterative optimization-based clustering methods (fcm) and model identification methods (like anfis). The subclust function finds the clusters by using the subtractive clustering method.
The genfis2 function builds upon the subclust function to provide a fast, one-pass method to take input-output training data and generate a Sugeno-type fuzzy inference system that models the data behavior.
An Example: Suburban Commuting
In this example we apply the genfis2 function to model the relationship between the number of automobile trips generated from an area and the area's demographics. Demographic and trip data are from 100 traffic analysis zones in New Castle County, Delaware. Five demographic factors are considered: population, number of dwelling units, vehicle ownership, median household income, and total employment. Hence the model has five input variables and one output variable.
tripdata creates several variables in the workspace. Of the original 100 data points, we will use 75 data points as training data (datin and datout) and 25 data points as checking data, (as well as for test data to validate the model). The checking data input/output pairs are denoted by chkdatin and chkdatout. The genfis2 function generates a model from data using clustering, and requires you to specify a cluster radius. The cluster radius indicates the range of influence of a cluster when you consider the data space as a unit hypercube. Specifying a small cluster radius will usually yield many small clusters in the data, (resulting in many rules). Specifying a large cluster radius will usually yield a few large clusters in the data, (resulting in fewer rules). The cluster radius is specified as the third argument of genfis2. Here we call the genfis2 function using a cluster radius of 0.5.
genfis2 is a fast, one-pass method that does not perform any iterative optimization. An FIS structure is returned; the model type for the FIS structure is a first order Sugeno model with three rules. We can use evalfis to verify the model.
The variable trnRMSE is the root mean square error of the system generated by the training data. To validate the model, we apply test data to the FIS. For this example, we use the checking data for both checking and testing the FIS parameters.
chkfuzout=evalfis(chkdatin,fismat); chkRMSE=norm(chkfuzout-chkdatout)/sqrt(length(chkfuzout)) chkRMSE = 0.6170
Not surprisingly, the model doesn't do quite as good a job on the testing data. A plot of the testing data reveals the difference.
At this point, we can use the optimization capability of anfis to improve the model. First, we will try using a relatively short anfis training (50 epochs) without implementing the checking data option, but test the resulting FIS model against the test data. The command-line version of this is as follows.
After the training is done, we type
fuzout2=evalfis(datin,fismat2); trnRMSE2=norm(fuzout2-datout)/sqrt(length(fuzout2)) trnRMSE2 = 0.3407 chkfuzout2=evalfis(chkdatin,fismat2); chkRMSE2=norm(chkfuzout2-chkdatout)/sqrt(length(chkfuzout2)) chkRMSE2 = 0.5827
The model has improved a lot with respect to the training data, but only a little with respect to the checking data. Here is a plot of the improved testing data.
Here we see that genfis2 can be used as a stand-alone, fast method for generating a fuzzy model from data, or as a pre-processor to anfis for determining the initial rules. An important advantage of using a clustering method to find rules is that the resultant rules are more tailored to the input data than they are in an FIS generated without clustering. This reduces the problem of combinatorial explosion of rules when the input data has a high dimension (the dreaded curse of dimensionality).
Overfitting
Now let's consider what happens if we carry out a longer (200 epoch) training of this system using anfis, including its checking data option.
[fismat3,trnErr,stepSize,fismat4,chkErr]= ... anfis([datin datout],fismat2,[200 0 0.1],[], ... [chkdatin chkdatout]);
The long list of output arguments returns a history of the step-sizes, the RMSE versus the training data, and the RMSE versus the checking data associated with each training epoch.
ANFIS training completed at epoch 200. Minimal training RMSE = 0.326566 Minimal checking RMSE = 0.582545
This looks good. The error with the training data is the lowest we've seen, and the error with the checking data is also lower than before, though not by much. This suggests that maybe we had gotten about as close as possible with this system already. Maybe we have even gone so far as to overfit the system to the training data. Overfitting occurs when we fit the fuzzy system to the training data so well that it no longer does a very good job of fitting the checking data. The result is a loss of generality. A look at the error history against both the training data and the checking data reveals much.
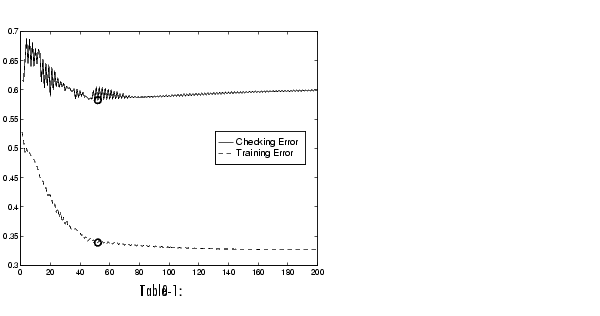
Here we can see that the training error settles at about the 50th epoch point. In fact, the smallest value of the checking data error occurs at epoch 52, after which it increases slightly, even as anfis continues to minimize the error against the training data all the way to epoch 200. Depending on the specified error tolerance, this plot also indicates the model's ability to generalize the test data.
A Clustering GUI Tool
There is also the Clustering GUI, which implements fcm and subclust, along with all of their options. Its use is fairly self-evident.
The clustering GUI looks like this, and is invoked using the command line function, findcluster.
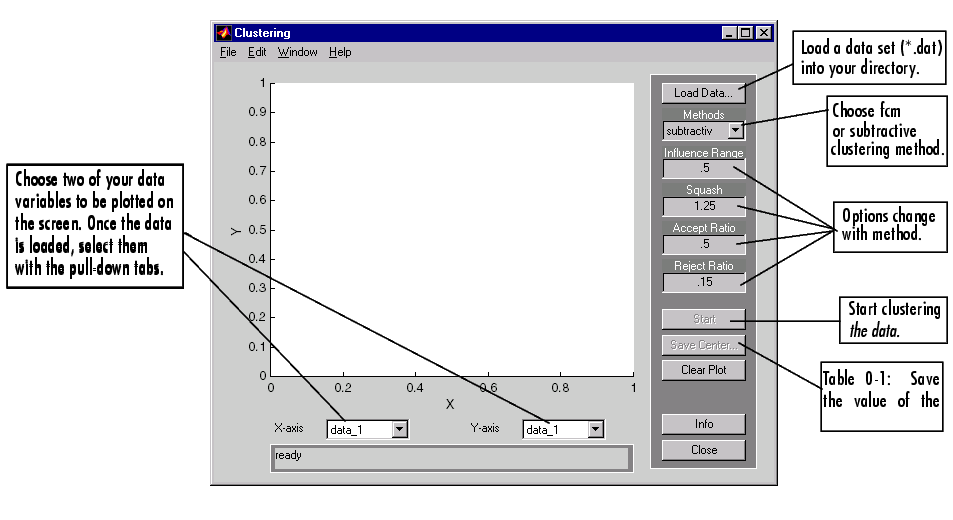
You can invoke findcluster with a data set directly, in order to open the GUI with a data set. The data set must have the extension .dat. For example, to load the data set, clusterdemo.dat, type findcluster('clusterdemo.dat').
You use the pull-down tab under Method to change between fcm (fuzzy c-means) and subtractive (subtractive clustering). More information on the options can be found in the entries for fcm, and subclust, respectively.
The Clustering GUI works on multidimensional data sets, but only displays two of those dimensions. Use the pull-down tabs under X-axis and Y-axis to select which data dimension you want to view.
 | Fuzzy C-Means Clustering | Stand-Alone C-Code Fuzzy Inference Engine |  |